Hartmut Michel – Molecular Membrane Biology
How to generate and use structural information to investigate function and mechanisms of membrane protein complexes?
Basic features of biological membranes and the function of membrane proteins.
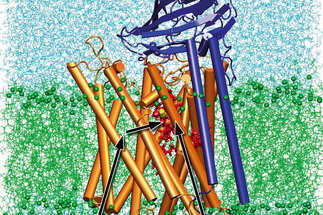
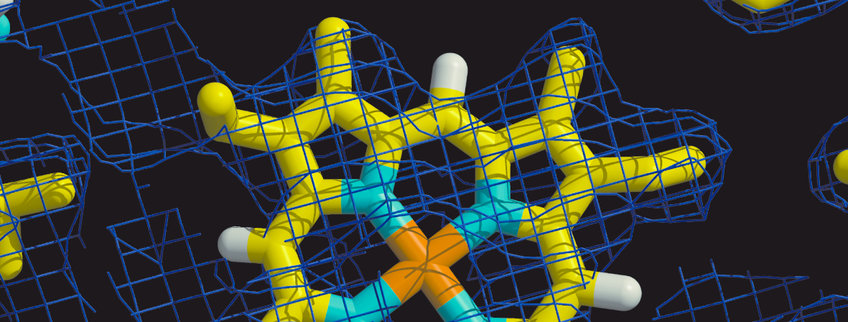
Membranes surround biological cells and divide the cells of higher organisms into various compartments. Biological membranes are composed of lipids, which form a bilayer, and of membrane proteins which are incorporated into the lipid bilayer. Lipid bilayers are impermeable to ions and to polar substances. As a consequence of this property ion gradients and electric voltages (“membrane potentials”) can be formed across membranes. Membrane proteins are required to enable the specific passage or transport of selected substances across membranes. Enabling passage and transport is therefore one of the most important functions of membrane proteins. Cells have to communicate, receive signals and to sense their environment. Many membrane proteins are sensors and receptors, the signal is often received at the outer face of the membrane and transduced across the membrane. Membrane proteins are central components of biological energy conversion. In photosynthesis and cellular respiration the transport of electrons and protons across membranes is the primary step of energy conversion. The resulting membrane potentials and ion gradients can be used to drive the synthesis of adenosine-5’-triphosphate (ATP), the uptake of nutrients, the export of waste products and proteins and flagella-based cell motility. Finally, some membrane proteins are enzymes, in particular, when the substrates and/or products are hydrophobic.
Summarizing, the functions of membrane proteins can be classified as follows:
- Passage and transport. Passage of substances and ions (“passive transport”) is catalysed by channels, pores and permeases. “Primary active transport” is driven by ATP or other energy-rich substances. In “secondary active transport” the downhill flow of an ion (normally a sodium ion or a proton) is coupled to the transport of a substrate.
- Signal reception and transduction. The most important receptors are the G-protein coupled receptors. Binding of an agonist to the receptor leads to an activation of trimeric G-proteins and the start of a signal transduction cascade.
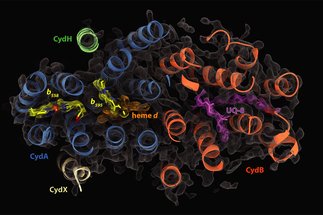
- Biological energy conversion. The prime examples are the respiratory chains of mitochondria and bacteria, where oxidation of substrates is coupled to the transfer of electric charges across the mitochondrial or bacterial membrane. In photosynthesis the photosynthetic reaction centres use the energy of light for the primary charge separation and transport of electrons across the photosynthetic membrane.
- Enzymes, preferentially for hydrophobic substrates and/or products.
The functional classification of a membrane protein may not be unique, because a receptor might be a channel protein working by opening or closing a channel upon interaction with its ligand.