Bonnie Murphy – Redox and Metalloproteins
Understanding mechanism through structure
We are an independent Max Planck Research Group based at the Max Planck Institute of Biophysics in Frankfurt. We work to discover the structure and mechanism of proteins, mostly proteins of bioenergetic relevance, using single-particle cryo-EM paired with biochemical, molecular and electrochemical techniques.
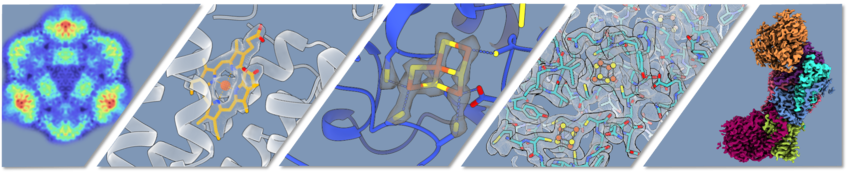
Our mission is to understand, at an atomic level, the function of sophisticated proteins that mediate the chemistry of life. Our focus is on metalloproteins, especially redox proteins, that play essential roles in shaping our planet, our climate, and our health. Most projects in the group use single-particle cryo-EM, a versatile and powerful tool that allows us to determine structures of protein complexes at atomic or near-atomic resolutions, without the need to grow crystals. At least as important as the static structure are structural changes in the complex, often linked to changes in its redox state, which mediate protein function. Due to the relatively flexible nature of cryo-EM sample preparation, combined with an ability to separate mixed populations of different conformational states in silico, single-particle analysis gives us an unprecedented ability to understand mechanism through the lens of structure.
Our work is built upon a foundation of biochemical and molecular biology techniques. In order to move beyond static structures to mechanistic storylines, we are creative with our approach to sample preparation, borrowing equipment and methods from other fields. This has allowed us to study oxygen-sensitive proteins, stably reduced states, and redox reactions under turnover on the grid.
Many projects apply single-particle cryo-EM to answer specific biological questions. In addition to this, we also work to expand the toolkit of cryo-EM. Currently, interpretation of densities due to bound metals and other ions, lipids, substrates and inhibitors entails substantial ambiguity, and we see a need for a method to sensitively map elemental distributions in cryo-preserved samples. By combining techniques from analytical electron microscopy with low-dose, automated data collection and image processing tools from single-particle analysis, we are developing a method for elemental mapping in dose-sensitive, cryo-preserved samples.
Our group is diverse and highly interdisciplinary and we welcome applications from candidates specialising in a wide range of fields including microbiology, biochemistry, chemistry and physics. Candidates interested in working in the group should send a CV and a letter of motivation to Bonnie Murphy.
Structures of a Methanogenic Megacomplex Shed Light on Conformationally-Gated Electron Transfer
Selected Publications and Preprints
