ATP synthases
Project Group of the Structural Biology Department
< go back to other projects
Membrane proteins and membrane protein complexes are more of a challenge for cryoEM than soluble complexes, partly because they are less rigid, but also because they are less stable and more difficult to prepare. Mitochondrial ATP synthases have been a long-standing research interest in the Department. ATP synthases produce virtually all the ATP in the cell by rotary catalysis. A trans-membrane proton gradient drives a rotor in the membrane, which powers ATP generation. The exact mechanism by which this happens remains elusive. We have made significant progress towards fully elucidating this strictly conserved process, which is one of the most fundamental and ancient in biology.
All types of rotary ATPases conform to essentially the same building plan. F-type ATPases consist of a roughly globular, water-soluble F1 head and the Fo subcomplex in the membrane. The F1 head contains three catalytic β and three non-catalytic α subunits that alternate in a hexameric (αβ)3 ring. The Fo subcomplex consists of the c-ring rotor, the stator subunits a and b, and, in mitochondria, a number of other small hydrophobic subunits. V-type and A-type ATPases correspondingly have a globular V1 (A1) catalytic head and a Vo (Ao) subcomplex with a rotor ring in the membrane. In all rotary ATPases, the central stalk links the catalytic head to the rotor ring. The rotor rings of F-type ATP synthases have 8 to 15 c-subunits (Figure 2). Most c-subunits are hairpins of two hydrophobic trans-membrane helices, whereas the ring subunits of V- and A-type ATPases often have two or more trans-membrane helix hairpins. In addition to the central stalk, F-type ATPases have one peripheral stalk, which connects the catalytic head to the stator in the membrane to prevent idle rotation of the whole assembly. The A-type ATPases have two peripheral stalks and V-type ATPases three.
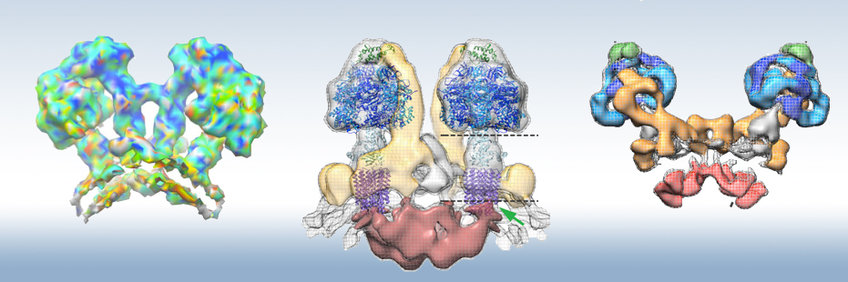
(a) Mitochondrial ATP synthase from Polytomella (Werner Kühlbrandt)
We determined the structure of the 1.6 MDa ATP synthase dimer from mitochondria of the chlorophyll-less green alga Polytomella sp. by single-particle cryoEM at 2.7 Å resolution. The Polytomella dimer is more rigid than that of yeasts (see below) or mammals and lends itself better to high-resolution cryoEM. The structure shows the arrangement of the two catalytic F1 heads, the central and peripheral stalks, and a pair of rotor rings in the dimer complex. We discovered that the a-subunit forms a bundle of four long membrane-intrinsic α-helices at right angles to the c-ring rotor. The long horizontal helices create a pair of proton half-channels at the a/c interface that enables protonation of the c-subunit glutamates from the lumenal side and proton release to the matrix, driving c-ring rotation and ATP synthesis (Allegretti et al., Nature 2015). Since our discovery, the long, quasi-horizontal helix hairpin of the a-subunit next to the c-ring rotor and the two half channels have proven to be fundamental, highly conserved feature of all rotary ATPases (Kühlbrandt and Davies, TIBS 2016)
(b) Mitochondrial ATP synthase from Yarrowia lipolytica (Werner Kühlbrandt, Thomas Meier)
ATP synthase dimers from yeasts resemble those from mammalian mitochondria but differ from those of plants and green algae in terms of structure and subunit composition. To gain insight into this more common and medically relevant ATP synthase, we performed single-particle cryoEM on dimers from the aerobic yeast Y. lipolytica, which we found to be more suitable for high-resolution cryoEM than the bovine complex. The Y. lipolytica dimer is an assembly of 17 different polypeptides in 62 individual protein subunits. The 6.2 Åresolution cryoEM map indicates a sharp ~90° bend in the detergent micelle surrounding the Fo part, reflecting the high local curvature at the ridges of the inner membrane cristae (Hahn et al., Mol Cell 2016). In the membrane region of the complex, we resolved and identified seven different membrane subunits, including the dimer-specific subunits e and g, which we have shown previously to be essential for dimer stability and cristae formation (Davies et al., PNAS 2012).
(c) Electron cryo-tomography of mitochondrial ATP synthases in situ (Werner Kühlbrandt)
Unlike the bacterial and chloroplast ATP synthase, or the V and A-type ATPases, all known mitochondrial ATP synthases form dimers in the membrane. as we discovered by cryoET of intact mitochondria or cristae vesicles (Strauss et al., EMBO J 2008; Davies et al., PNAS 2011; Daum et al., PNAS 2013). The dimers arrange in long rows along the tightly curved ridges of the mitochondrial inner membrane cristae. The dimer rows have a profound effect on respiratory activity, growth rates and mitochondrial morphology. The rows are instrumental for the formation of the inner membrane cristae, which increase the membrane area available for respiratory chain complexes. The cristae lumen can be considered as a mitochondrial sub-compartment that may assist ATP production through local variations in proton concentration.
Cristae vesicles from protozoan mitochondria often look very different from the standard morphology. Subtomogram averaging revealed that the dimers themselves can be very different, especially with respect to the angle included by the central stalks, which is ~90° in fungi and mammals but ranges from around 60° (trypanosomes, Polytomella) to 0° (Paramecium). Differences are striking in Paramecium, where U-shaped ATP synthase dimers assemble into regular helical arrays, which impose a helical structure and thereby membrane curvature on the tubular cristae that are characteristic of this and similar species. Remarkably, the dimer rows induce high, uniform membrane curvature, even though the dimer angle is 0° and therefore an individual dimer does not bend the membrane (Mühleip et al., PNAS 2016b). Differences are even more striking in trypanosomes and Euglena, where the structure of the F1 subcomplex that was thought to be universally conserved in all F-type ATPases deviates from the consensus structure. We discovered that in the Euglenacea, which include dangerous human parasites, the α and β-subunits form a trigonal pyramid , rather than the usual near-sixfold barrel. This unusual architecture is due to a split in the α-subunit into separate N and C-terminal domains, which disrupts the catalytic site of ATP production in the F1 head. The unique structure of these ATP synthases, on which the organisms depend for survival, opens a new route for drug development to combat sleeping sickness that is caused by trypanosomes (Mühleip et al., PNAS 2016a).


