Membrane transporters, channels and enzymes
Project Group of Structural Biology Department
< go back to other projects
Membrane transporters, channels and enzymes
The first structure of a secondary-active transporter, the E. coli sodium/proton antiporter NhaA was determined at 6 Å resolution by electron crystallography in this Department (Williams, Nature 2000; Williams et al., EMBO J 1999). Since then, numerous x-ray structures of secondary-active transporters have been published by us and others. With recent developments, even comparably small membrane transporters and ion channels are now within reach of single-particle cryoEM.
Crystallography of sodium-proton antiporters
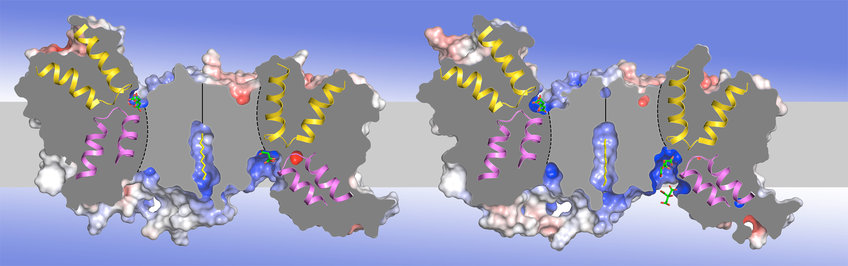
The electroneutral Na+/H+ antiporter NhaP from the hyperthermophilic archaeon Pyrococcus abyssi (PaNhaP) is a functional homologue of the medically important human antiporter NHE1. We determined the structure of PaNhaP by x-ray crystallography at pH 8 (Figure 1A) and pH 4. The substrate ion was resolved for the first time in such a transporter by soaking crystals with thallium, an anomalous x-ray scatterer that mimics Na+ in the binding site (Figure 1B). Transport measurements show that thallium is a substrate for PaNhaP. The substrate ion is coordinated by three acidic sidechains, a water molecule, a serine and a main-chain carbonyl in the unwound stretch of TM helix 5 (Wöhlert et al., eLife 2014).
We also determined the structure of the related Na+/H+ antiporter NhaP1 from Methanococcus jannaschii (MjNhaP1) in two complementary states. The 3.5 Å x-ray structure of the inward-open state in the presence of sodium at pH 8, where the transporter is highly active, was solved by molecular replacement with PaNhaP. The 6 Å structure of the complementary outward-open state was obtained by electron crystallography of 2D crystals at pH 4, where the transporter is inactive (Paulino et al., eLife 2014). Comparison revealed a 7° tilt of the 6‑helix bundle that includes the ion-binding site. Progressive substrate-induced conformational changes in MjNhaP1 were investigated by difference maps obtained from 2D crystals incubated with a range of physiological pH and Na+ conditions (Paulino and Kühlbrandt, eLife 2014). Changes in Na+ concentration caused a marked conformational change that was largely pH-independent, consistent with substrate competition for a common ion-binding site, as confirmed by electrophysiological studies in collaboration with Klaus Fendler in the Bamberg department (Calinescu et al., J Biol Chem 2014). Projection difference maps indicated helix movements that convert MjNhaP1from the proton-bound, outward-open state to the Na+-bound, inward-open state. Oscillation between the two states results in rapid antiport.
The citrate transporter SeCitS
We solved the x-ray structure of the sodium/citrate symporter SeCitS from the human pathogen Salmonella enterica, which takes up citrate as a nutrient. Unexpectedly, we found that the structure of bacterial citrate transporters closely resembles that of the sodium/proton antiporters, without detectable sequence homology. The 2.5 Å resolution x-ray structure of SeCitS contains two dimers. In each dimer, the two protomers assume different, inside-open or outside-open conformations that do not result from crystal contacts. The structure thus shows three functional states of the active protomer in one asymmetric unit (Figure 1C-E), which is unprecedented. Together with comprehensive functional studies of SeCitS reconstituted into liposomes, the structures explain the transport mechanism as a six-step process, with a rigid-body 31° rotation of a helix bundle around an axis in the membrane plane, perpendicular to the long dimer axis. The rotation of the helix bundle translocates the bound substrates vertically by 16 Å across the membrane. We expect similar transport mechanisms apply to a wide range of secondary transporters (Wöhlert et al., eLife 2015).
Figure 1. X-ray structures of secondary-active transporters

CryoEM of transporters and ion channels
The human TRP channel Polycystin-2 (PC2) responds to changes in cytosolic calcium concentration. PC2 mutations trigger autosomal dominant polycystic kidney disease. PC2 is found predominately in the endoplasmic reticulum (ER), in the primary cilium and, depending on cell type, in the plasma membrane. We determined cryoEM structures of two full-length PC2 channels from the plasma membrane or the endoplasmic reticulum at 4.2 Å resolution (Figure 2). The structures show two distinct open states in complex with lipids and Ca2+. A single Ca2+ is bound in the selectivity filter, while multiple Ca2+ ions along the translocation pathway suggest an electrostatic knock-off mechanism to increase Ca2+-selectivity outside the ER (Wilkes et al., NSMB 2016).
PilQ, a bacterial protein export/DNA uptake system
Bacteria use flagella or pili for locomotion. While flagellar motors are well-characterized, much less is known about the machinery that generates bacterial pili. The type 4 pilus (T4P) secretion complex from Thermus thermophilus is a 1100 kDa assembly of several different membrane and membrane-associated proteins. The T4P complex is active both in the extrusion of pili and in the uptake of DNA during bacterial transformation. In collaboration with Beate Averhoff (Frankfurt University) we are determining the structure of the T4P complex by cryoEM. Subtomogram averages of T4P in entire Thermus thermophilus cells show the complex in the closed and open state, with a pilus in the process of being extruded (Gold et al., eLife 2015).

