Bacterial pore-forming toxins
Project Group of Structural Biology Department
< go back to other projects
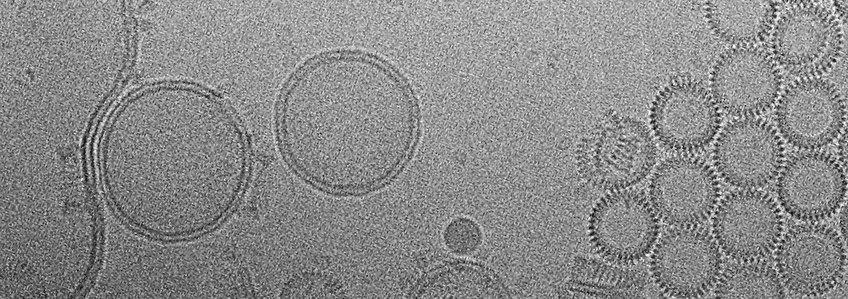
Gram-positive pathogens, such as Pneumococcus, Listeria and Clostridium, secrete cholesterol-dependent cytolysins (CDCs) as soluble monomers. The monomers attach to cholesterol-containing target membranes, where they assemble into large, circular pores that perforate the target cells. The structure and mechanism of pore-forming toxins are studied in the project group of Dr. Özkan Yildiz in the Department, who determined the x-ray structures of two CDCs in the soluble form: listeriolysin (LLO) from Listeria monocytogenes (Köster et al., Nat Comm 2014), and pneumolysin (PLY) from Streptococcus pneumoniae (van Pee et al., Nano Letters 2016). To understand the mechanism of pore formation and membrane perforation indetail (Video 1), we determined the structure of the ~2.2 MDa pore complex of PLY by single-particle cryoEM at a resolution of 4.5 Å (Figure 1). The PLY pore complex is a 400-Å ring of 42 membrane-inserted monomers. During membrane insertion, one of the four protein domains in the soluble monomer refolds into two ~85 Å β‑hairpins that traverse the lipid bilayer. In the membrane, the hairpins assemble into a cylindrical 168-stranded β‑barrel (Figure 1A,B insets). Large sidechains are resolved on the apolar outer barrel surface, while the inner barrel surface is highly charged (Figure 1B). CryoET shows that the soluble monomers assemble on cholesterol-containing liposomes into multi-subunit, circular prepores, with residues that confer lipid specificity on the membrane surface. Tomographic volumes show that the lipid bilayer in the prepore is intact (Figure 1C,D), while it has disappeared in the mature pore (Figure 1E,F). Presumably the highly charged inner pore surface repels and helps to disperse any lipid trapped in the pore (van Pee et al., eLife 2017).
