High-resolution cryoEM of soluble multi-protein complexes
Research by Janet Vonck & Werner Kühlbrandt
< go back to other projects
Near-atomic (sidechain) resolution by cryoEM is achieved most readily with stable, water-soluble complexes. Although our main focus is on membrane proteins and membrane systems, we have determined the structures of three such soluble macromolecular assemblies, to establish high-resolution techniques in the Department and to find out how far we can push them.
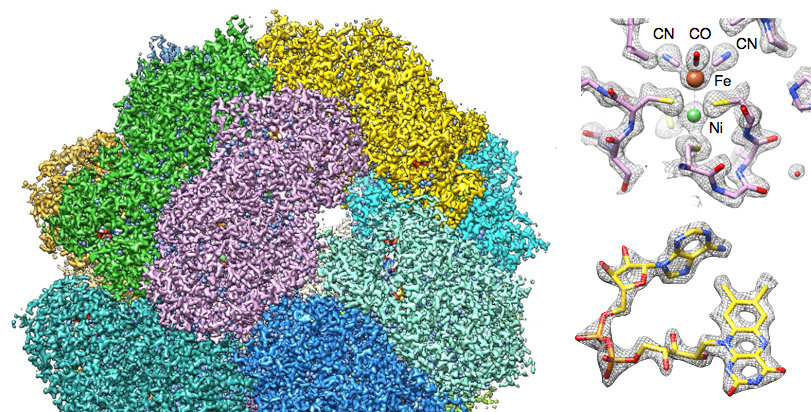
As our first near-atomic resolution cryoEM structure, and one of the very first in the “resolution revolution”, we obtained a 3.4 Å map of the 1.2 MDa multi-enzyme hydrogen transferase Frh (Allegretti et al., eLife 2014) (Figure 1A). The density map (Figure 1B) is very clear, due to the high quality of experimentally determined phases, making it easy to fit the polypeptide chains and prosthetic groups with standard crystallographic programs. By re-processing the published image data and adding new, high-quality images, we have increased the resolution significantly and just broke the 3 Å boundary for the first time. Another soluble protein complex we work on is the alcohol oxidase from Pichia pastoris, which had defeated x-ray crystallography for decades. Figure 1C shows its cryoEM structure at 3.4 Å resolution (Vonck et al., PLoS One 2016).
